Search Results for 'Sequences Genome'
Sequences Genome published presentations and documents on DocSlides.
Biomonitoring, but not as we know it:
meeting the challenge of DNA-based observation in...
ML Approaches – Conceptual Stuff
Nitin Kohli. DS W210 – Capstone Project. Sequen...
Global Variation in Copy Number
in the Human Genome. Redon et. al.. Presentation ...
Genetic
“. Engineering. ”. for Crop Improvement. Xiw...
Haploid-Diploid Evolutionary Algorithms
Larry Bull. UWE. SEX. . A Social Interaction in ...
Neutral theory:
The vast majority of observed sequence difference...
Introduction to Short Read Sequencing Analysis
Jim Noonan. GENE 760. Sequence read lengths remai...
Cufflinks
Matt . Paisner. , . Hua. He, Steve Smith and Bri...
3D Genome Workshop - Overview of ENCODE
Dan Gilchrist. National Human Genome Research Ins...
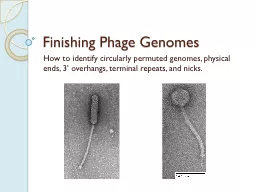
Finishing Phage Genomes
How to identify circularly permuted genomes, phys...
http://cs273a.stanford.edu [BejeranoFall14/15]
1. MW . 12:50-2:05pm . in Beckman . B100. Profs...
The History of Genetics
From classical genetics to the personal human gen...
http://cs273a.stanford.edu [Bejerano Fall16/17]
1. CS273A. Lecture . 14: . Inferring Evolution: C...
Introduction To Next Generation Sequencing (NGS) Data Anal
Jenny Wu. UCI Genomics High Throughput Facility. ...
Ensembles of HMMs and their use in
biomolecular. sequence analysis. Nam-phuong Nguy...
FREQUENT PATTERNS IN
CALIFORNIA . COMMUNITY COLLEGE. STUDENT COURSE SE...
Adversarial Learning for Neural Dialogue Generation
2017.2.17. Zhang Yan. 1. Li. ,...
Transcriptomics
Jim Noonan. GENE 760. Transcriptomics. Introducti...
Joseph Loquasto
Department . of Food Science. The Pennsylvania St...
Genomes and their evolution
Chapter 21. You must know. How prokaryotic genome...
Organelle genomes
Organelle gene expression processes. Organelle-to...
Wrapup
NHGRI strategic plan. What does the NIH think gen...
Genome sequencing and assembly
Mayo/UIUC Summer . C. ourse in Computational Biol...
John Sherwood
1. , Victoria Torres. 1. , Jasmina Cunmulaj. 1. ,...
MCB 3421 class 25
s. tudent evaluations. Please follow this link to...
A high-resolution map of human evolutionary constraint usin
Kerstin Lindblad-Toh1 et al.. Presentation by:. K...
Dot plot
Daniel Svozil. Software choice. source: Bioinform...
Marker heritability
Biases, confounding factors, current methods, and...
MAFFT:
Multiple Sequence Alignment using Fast Fourier Tr...
Genomics
interventions towards fast-track development of ....
Algenol’s proprietary enhanced algae, AB1, are
non-invasive. to local SWFL waters . and get out...
Phycodnaviridae
Abigail S. Clark. MCB 5505: General Virology. Pho...
Constructing the Tree of Life: Divide-and-Conquer!
Tandy Warnow. University of Illinois at Urbana-Ch...
Database search
and . pairwise. alignments. 1. “. It is a capi...
Evaluating strategies for pandemic response in Delhi using
Huadong. Xia. Joint work with . Kalyani. . Nag...
Genome Sciences 373
Genome Informatics . Q. uiz Section . 9. May 26 2...
What’s New in Manageability for Microsoft SQL Server “D
DBI304. Denny Cherry. Sr. Database Administrator....
Copyright © 2024 DocSlides. All Rights Reserved











![http://cs273a.stanford.edu [BejeranoFall14/15]](https://thumbs.docslides.com/538561/http-cs273a-stanford-edu-bejeranofall14-15.jpg)

![http://cs273a.stanford.edu [Bejerano Fall16/17]](https://thumbs.docslides.com/540408/http-cs273a-stanford-edu-bejerano-fall16-17-.jpg)