Search Results for 'Transcriptome Seq'
Transcriptome Seq published presentations and documents on DocSlides.
Frédéric CHOULET A pseudomolecule of 774 Mb: the 3B experience
INRA GDEC – Clermont-Ferrand, France. 3B. Sequen...
Regulatory Genomics Lab Regulatory Genomics | Saurabh Sinha | 2021
1. Part 1: . ChIP. -seq peak calling. Part 2. Anal...
The Cancer Genome Atlas (TCGA) and
cBioPortal. . Nitish . Mishra. TCGA history. TCGA...
JAX: Exploring The Galaxy
Glen Beane, Senior Software Engineer. The Jackson ...
Measuring TCP RTT in the
. data. . plane. Xiaoqi. Chen. , . Hyojoon. Kim...
APPLICATION FOR REGISTRATION OF OR CIGAR LOUNGEContinuedOC53MAY 07
14.Mailing Address of Registered Agent for Busines...
PAGE0003 TITLE
SEQ 0003 JOB 0094X-002-008 18DEC03 AT 09:OO D...
IntroductiontoMicroarrayAnalysisA11ymetrixGeneChiptechnologyKaterin
IntroductiontoMicroarrays{Goal GoalofthetalkIRevie...
genomeannotationbasedonclusteringfixedlengthgenomiclocithatcanbemodel
boundariesareenforcedtohavethesamegenomicpos-ition...
ELSEVIERAbstractKeywordsElectron beam tomography Spiral computed tom
182[9-13].Recent studies have documented the abili...
initiatedbytheNeurosporahomologueofGCN5NGF1whichacetylateshistoneH
intranscriptdiscovery.Intotal,weidentified21,475is...
Cell type identification in single-cell RNA-
seq. data. Philip Lijnzaad. Overview. Introductio...
The R Language Dr. Smruti
R. Sarangi and Ms. . Hameedah. Sultan. Computer S...
Figure Figure. . Neighbor-joining tree of Coxiella burnetii genotypes determined by multispacer seq
Angelakis E, Johani S, Ahsan A, Memish Z, Raoult D...
Py rope Towards provably correct hardware modeling in Python/
HeteroCL. Presenter: . Deyuan (Mike) He, Universit...
Introduction to Epigenetics
BMI/CS 776 . www.biostat.wisc.edu/bmi776/. Spring ...
Transport Layer 3- 1 Chapter 3
Transport Layer. Computer Networking: A Top Down A...
Transport Layer 3- 1 Chapter 3 outline
3.1 transport-layer services. 3.2 multiplexing and...
Transport Layer CS 3516 – Computer Networks
Chapter 3: Transport Layer. Goals:. Understand . p...
Computational Methods
for A. nalysis of Single . C. ell RNA-. Seq. Data...
Data Analysis in Next Generation
Sequencing. . Paolo Aretini . Senior . Researcher...
Considerations on LTF Sequence Design
Date:. . 2015-05-10. Authors:. LTF. -related deci...
Cinderella: Turning Shabby X.509 Certificates into Elegant Anonymous Credentials
Antoine Delignat-Lavaud. Cédric Fournet, Markulf ...
Biostatistics & Bioinformatics Shared Facility
Sejong Bae, PhD, FMSSANZ, . Young-il Kim, PhD . El...
Factors shaping structural organization of chromatin
Ekaterina . Khrameeva. Skolkovo. Institute of Sci...
Product Strategy for truChIP® and cryoPREP®
Sean Gerrin, ALM . Product Manager, Epigenomics. w...
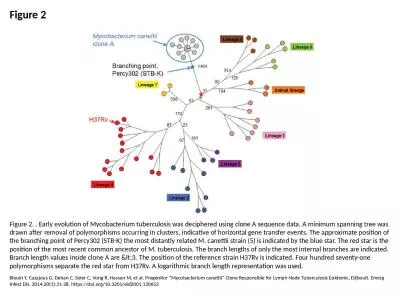
Figure 2 Figure 2. . Early evolution of Mycobacterium tuberculosis was deciphered using clone A seq
Blouin Y, Cazajous G, Dehan C, Soler C, Vong R, Ha...
CORTEX seq-FISH: gene selection
Hang Xu. Stanford University. seqFish. & . sc...
Regulatory Genomics Lab Regulatory Genomics | Saurabh Sinha | 2023
1. Part 1: . ChIP. -seq peak calling. Part 2. Anal...
Cankun Wang Department of Agronomy, Horticulture & Plant Science
7/17/2019. Development . of Computational . Techni...
April 20, 2017 Jerome I. Rotter, MD and Stephen S. Rich, PhD
MESA Steering Committee. Genetics Committee Update...
DNS, Web, TCP Sequence Numbers
EE122 Discussion. 10/19/2011. DNS. Mapping between...
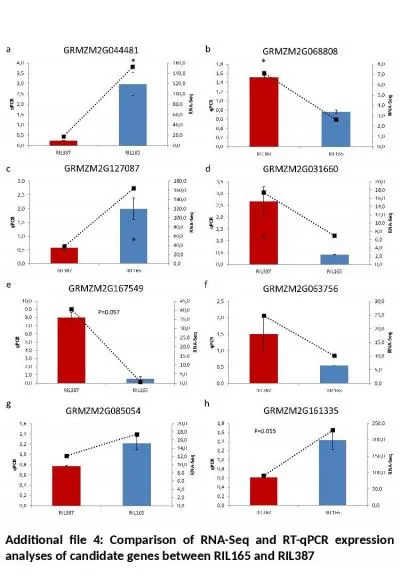
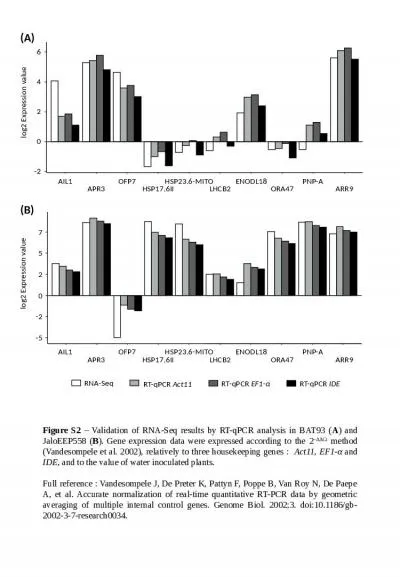
Additional file 4: Comparison of RNA-
Seq. and RT-qPCR expression analyses of candidate...
RNA-Seq RT-qPCR Act11 RT-qPCR
EF1-. α. RT-qPCR . IDE. -2. 0. 2. 4. 6. AIL1. APR...
Docexpress – A galaxy
docker. for estimation of Expression profiling in...