PPT-Identifying novel sequence variants of RNA 3D motifs
Author : danya | Published Date : 2024-01-13
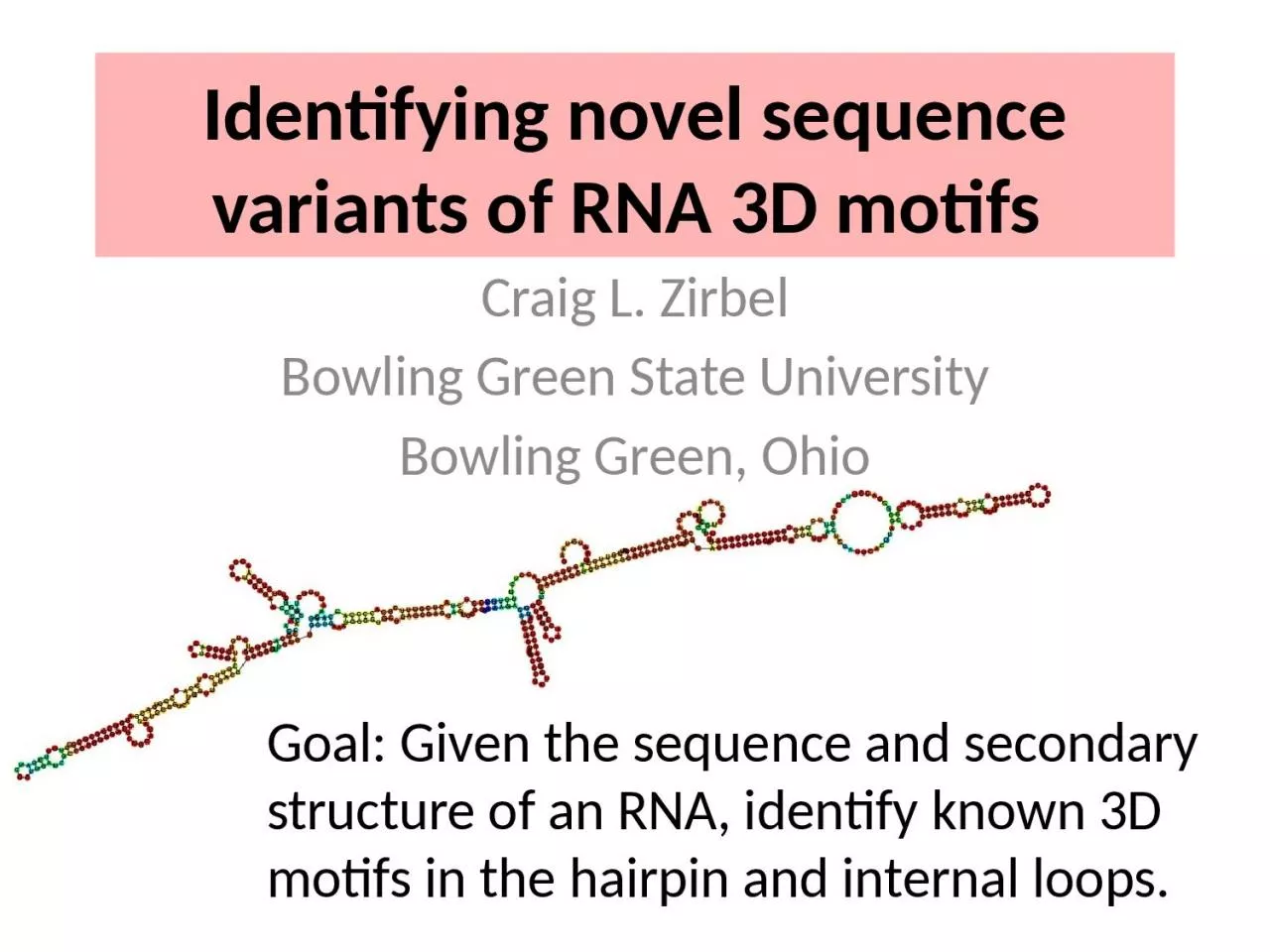
Goal Given the sequence and secondary structure of an RNA identify known 3D motifs in the hairpin and internal loops Craig L Zirbel Bowling Green State University
Presentation Embed Code
Download Presentation
Download Presentation The PPT/PDF document "Identifying novel sequence variants of ..." is the property of its rightful owner. Permission is granted to download and print the materials on this website for personal, non-commercial use only, and to display it on your personal computer provided you do not modify the materials and that you retain all copyright notices contained in the materials. By downloading content from our website, you accept the terms of this agreement.
Identifying novel sequence variants of RNA 3D motifs: Transcript
Download Rules Of Document
"Identifying novel sequence variants of RNA 3D motifs"The content belongs to its owner. You may download and print it for personal use, without modification, and keep all copyright notices. By downloading, you agree to these terms.
Related Documents